Replication timing using transient hemimethylation
Kathy Friedman's protocol; see Friedman, K. L., M. K. Raghuraman, W. L. Fangman, and B. J. Brewer (1995) Analysis of the temporal program of replication initiation in yeast chromosomes. J. Cell Sci. Suppl. 19:51-58.
Rationale of methylation timing approach
The E. coli dam methylase adds methyl groups to adenine in the sequence 5' GATC 3' ( Marinus and Morris, 1973; Geier and Modrich, 1979 ). This enzyme methylates both unmethylated and hemi-methylated sites. In yeast DNA, methylated bases are undetectable, unlike the DNA of most higher eukaryotes ( Hattman et al., 1978 ). However, the dam methylase protein can be expressed at moderate levels in yeast with few obvious effects on cell viability or growth rate ( Brooks et al., 1983; Hoekstra and Malone, 1985 ). Expression of the protein results in moderate levels of DNA methylation. The accessibility of different GATC sites to methylation is variable, presumably due to differences in chromatin structure that affect the access of the enzyme to DNA ( Gottschling, 1992; Singh and Klar, 1992; Wright et al., 1992; Kladde and Simpson, 1994 ).
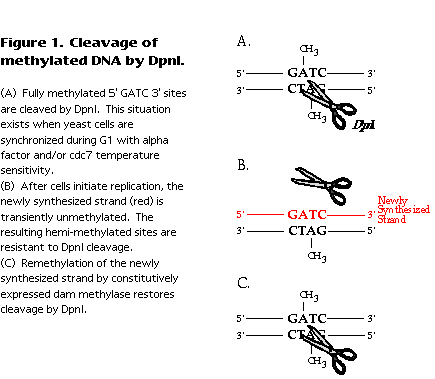
E. coli DNA, though normally fully methylated, undergoes a transient period of hemimethylation immediately following passage of the replication fork (reviewed in Barras and Marinus, 1989 ). The newly synthesized strand is not initially methylated, but acquires methylation after dam methylase contacts the newly replicated DNA sequence. Methylated and hemi-methylated sequences can be distinguished by their differential sensitivity to cleavage by DpnI (Geier and Modrich, 1979 ). Fully methylated DNA is cleaved at GATC sites and hemi-methylated or unmethylated DNA is not (Figure 1). Therefore, the time at which E. coli sequences become resistant to cleavage by DpnI correlates with their time of replication. A similar logic can be applied to replication of yeast DNA in a strain expressing the dam methylase. Again, replication fork passage results in hemi-methylation of the DNA, detectable as resistance to cleavage by DpnI (Figure 1).
Strain construction
In order to construct a strain containing constitutively methylated DNA, the E. coli dam methylase gene and its endogenous promoter were excised from plasmid pMFH1 ( Hoekstra and Malone, 1985 ) with HindIII and PvuII and inserted into the HindIII-SmaI sites of the polylinker in pRS305 (Sikorski and Hieter, 1989), a Leu+ yeast integrating vector. The resulting plasmid (pRS305-DAM) was partially digested with ClaI and the linearized plasmid was used to transform yeast strain RM14-3a (MATa cdc7-1 bar1 ura3-52 trp1-289 leu2-3,112 his6). Cleavage of transformant DNA with DpnI was assessed by Southern hybridization, and the transformant exhibiting the highest level of DpnI cleavage was selected (RM14-3a Meth5 strain).
Protocol for methylation timing
| 1. | Grow ~330 ml of RM14-3a expressing dam methylase (RM14-3a Meth5) to an OD660 of about 0.3 (3.85 x 106 cells/ml) at 23°C. Arrest with 200 nM alpha factor for 1-1.25 doubling times, until the percent budded cells stops increasing (usually ~95%). (RM14-3a is bar1 , so the alpha factor concentration required is low). |
| 2. | Transfer cells to 37°C and wait for culture itself to reach 37°C. Add Pronase (20 µg/ml final concentration -- dissolve Pronase in 5 ml minimal medium). We use Pronase from Calbiochem. |
| 3. | Hold at 37°C for ~120 minutes (until % budded cells reaches 95-98%). Remove two samples for 0 time point, then swirl the flask in ice water for one minute and return to 23°C. I usually collect samples every five minutes for 55 minutes, then collect 65 and 80 minute time points. |
| 4. | Collecting samples: Freeze 8 ml of 0.1% Na-azide, 0.2 M EDTA in "50 ml" Oakridge-type plastic screw cap centrifuge tubes (freeze these slanted at -20°C to increase surface area). During the experiment, mix 20 ml culture with a 1/20 volume of 10% Na-azide in a 35 ml Corex tube (on ice), immediately transfer to the tube of frozen EDTA /azide, and shake the tube to chill the sample and melt the frozen EDTA. Spin the cells down in a chilled centrifuge, wash with 1 ml cold water in Eppendorf tubes, and freeze the pellets at -20°C. |
| 5. | Extract the DNA by smash and grab method. |
| 6. | Restriction digest: |
| (a) We usually resuspend each DNA pellet in 20 microliters of TE, then digest half of each sample with EcoRI (this allows us to run two gels from one experiment). I typically use 1 microliter of EcoRI (at 20,000 units/ml) for each sample. After ~6 hours, the DNA is precipitated. One of the 0 minute time samples is saved at this point as a control for the size of the EcoRI fragment (i.e. don't cut one of the samples with DpnI). The rest of the samples are resuspended in 50 microliters water, and an equal volume of a digest mix containing DpnI, restriction buffer, and BSA is added. After ~6 hours, the DNA is precipitated and loaded in a 0.7% agarose gel. | |
| (b) After blotting the gel, the blot is sequentially hybridized with a fragment adjacent to ARS1 (early replicating), the R11 (late replicating) fragment, and any other fragments of interest. |
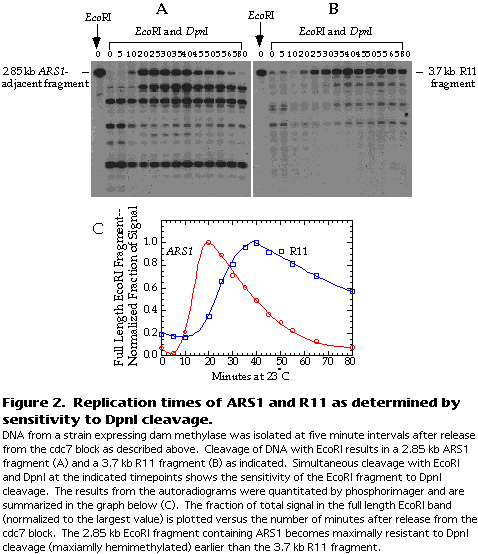
Figure 2A and B show a typical result for ARS1 and R11.
Quantitation
The blots are quantitated by phosphorimager. The extent of DpnI cleavage is expressed as the fraction of total signal in the lane that is attributable to the full-length EcoRI fragment. This fraction should increase as the fragment is replicated because the DNA becomes transiently resistant to DpnI cleavage. The fraction of signal in the full-length EcoRI fragment is then normalized to the largest value and plotted versus the time after release from the cdc7 block (see Figure 2C).
References
Barras, F., and M. G. Marinus. 1989. The great GATC: DNA methylation in E. coli. Trends Genet. 5: 139-143.
Brooks, J. E., R. M. Blumenthal, and T. R. Gingeras. 1983. The isolation and characterization of the Escherichia coli DNA adenine methylase (dam) gene. Nucleic Acids Res. 11: 837-851.
Geier, G. E., and P. Modrich. 1979. Recognition sequence of the dam methylase of Escherichia coli K12 and mode of cleavage of Dpn I endonuclease. J. Biol. Chem. 254: 1408-1413.
Gottschling, D. E. 1992. Telomere-proximal DNA in Saccharomyces cerevisiae is refractory to methyltransferase activity in vivo. Proc. Natl. Acad. Sci. U S A 89: 4062-4065.
Hattman, S., C. Kenny, L. Berger, and K. Pratt. 1978. Comparative study of DNA methylation in three unicellular eucaryotes. J. Bacteriol. 135: 1156-1157.
Hoekstra, M. F., and R. E. Malone. 1985. Expression of the Escherichia coli dam methylase in Saccharomyces cerevisiae: Effect of in vivo adenine methylation on genetic recombination and mutation. Mol. Cell. Biol. 5: 610-618.
Kladde, M. P., and R. T. Simpson. 1994. Positioned nucleosomes inhibit dam methylation in vivo. Proc. Natl. Acad. Sci. Usa 91: 1361-1365.
Marinus, M. G., and N. R. Morris. 1973. Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J. Bacteriol. 114: 1143-1150.
Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19-27.
Singh, J., and A. J. Klar. 1992. Active genes in budding yeast display enhanced in vivo accessibility to foreign DNA methylases: a novel in vivo probe for chromatin structure of yeast. Genes Dev. 6: 186-196.
Wright, J. H., D. E. Gottschling, and V. A. Zakian. 1992. Saccharomyces telomeres assume a non-nucleosomal chromatin structure. Genes Dev. 6: 197-210.
Return to top of page
